Incorporation of a Matrix Metalloproteinase-Sensitive Substrate into Self-Assembling Peptides – A M
- 格式:pdf
- 大小:418.40 KB
- 文档页数:7


CD38 是一个定位于膜上的糖蛋白,催化环腺苷二磷酸核糖(cADPR, cyclic ADP-ribose)的合成和降解。
cADPR 是核苷酸的代谢产物,通过作用于 ryanodine 受体( RyRs)参与细胞内钙库的钙动员。
许多研究发现 CD38/cADPR 介导的 Ca2+信号传递和通过 RyRs 通道的Ca2+释放在Ca2+内平衡的调控中发挥了重要的作用。
CD38/cADPR/ RyRs 介导的 Ca2+信号传递也参与了许多病理和生理过程。
本文就 CD38 基因的结构、表达、调控及在心血管系统的功能做一综述。
结构:cd38 基因包括了 8 个外显子,5′ UTR无 TATA盒或 CAAT盒( Nata K, Takamura T, Karasawa T, Kumagai T, Hashioka W, Tohgo A, Yonekura H, Takasawa S, Nakamura S, and OkamotoH. Human gene encoding CD38 (ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase):organization, nucleotide sequence and alternative splicing. Gene 186: 285– 292, 1997. )。
一段高 GC含量区可能是 cd38 的启动子区,调节 CD38的表达。
CD38 是一个 45kDa 的单链跨膜糖蛋白,整体结构分为N 末端短的胞质尾,单次跨膜域和 C 端长的胞外区(图 1)。
其开放阅读框含有300 个氨基酸残基, 5′UTR比较短小, 69bp;3′ UTR含有 260bp,不包括 PolyA 尾。
其编码的多肽链分子量为34, 288 道尔顿。
天然的CD38 抗原的分子量为46,000 道尔顿。
预测的氨基酸序列没有N- 末端的引导肽序列,但在从翻译起始位点起 21 个氨基酸残基处含有一段长23 个氨基酸残基的内部疏水序列。


Supplementary MethodsBiotinylation. Biotinylation of cell surface GLT1 was performed as described previously1, loading gels with 10mg protein from each of the following fractions: total cell homogenate, intracellular and membrane fractions. Mixed neuron/glial cu ltures were prepared from newborn mice (12-48 hours). The cortices were dissected from brain, incubated in trypsin for 15 min at 37º, repeatedly triturated, than plated onto poly-L-lysine (15µg/mL) coated plastic culture dishes. Cultures were maintained in DMEM (Invitrogen) containing 25mM of glucose, 19.4µM pyridoxine hydrochloride, 2mM L-glutamine, 5% fetal bovine serum (Invitrogen) and N2 supplement (Invitrogen). The cultures were confluent after 7 days when maintained in an incubator at 5% CO2 , 37ºC with biweekly media and drug changes. In some experiments, confluent cells were treated for 7 days with freshly prepared media containing ceftriaxone (100µM).Glutamate transporter currents. To provide an indication of the number of functional glutamate transporters present in astrocyte membranes, we made whole-cell recordings from astrocytes in area CA1 of hippocampal slices prepared from postnatal day (P) 17 to 19 rats, as described2, using an internal solution composed of: KNO3 130mM, EGTA 10mM, HEPES 20mM, MgCl2 1mM, Na2ATP 2mM, NaGTP 0.2 mM; the pH was 7.3. Transporter currents elicited in response to application of exogenous substrates activate only a fraction of available transporters, due to the high density of transporters in brain tissue3,4, presenting difficulties for measuring changes in transporter abundance. For this reason, we measured transporter associated anion currents from astrocytes. Anions such as NO3– permeate glutamate transporters in the absence of glutamate2,5, and induce a constitutive leak current. The amplitude of the substrate-independent current is proportional to the number of functional transporters and can be blocked by the antagonist DL-threo-β-benzyloxyaspartic acid (TBOA)2,6. For these experiments, astrocytes were voltage-clamped at –90 mV and the change in holding current produced by bath application of 200 M TBOA was measured. Animals were injected daily starting at P12 with 200 mg/ml i.p. ceftriaxone dissolved in isotonic saline or saline alone. Recordings were performed on slices from both ceftriaxone and saline injected animals on the same day; however, the treatment received by each rat was not revealed to the experimenter.Oxygen glucose deprivation/Ischemic preconditioning. In vitro ischemia was modeled by o xygen glucose deprivation (OGD). Cortical cells were either subjected to control treatment (media: modified Earle’s balanced salt solution including glucose, bubbled with 5% CO2 95% O2) without OGD performed, or 5 minutes (sublethal) of OGD (using modified Earle’s balanced salt solution which is devoid of glucose and bubbled with 10%H2, 85%N2, and 5% CO2 to deoxygenate). Anaerobic conditions were achieved using an anaerobic chamber at 37︒C. OGD was terminated by exchange of media back to oxygenated growth media. Twenty-four hours following the above, cortical cells were subjected either to no treatment or to one hour of OGD. Neuronal survival was determined by computer-assisted cell counting after staining with the fluorescent vital dyes propidium iodide (as an indicator of neuronal death) and Hoechst 33342 (as an indicator of total number of neurons) and is presented as percent of cell death. Glial nuclei fluoresce at a lower intensity and were gated out. Ceftriaxone (1 μM) was added 24 hours prior to the first experimental condition and thus was in the culture medium 48 hours prior to the onset of 1 hour OGD. Following 1 hour OGD, cells were returned to oxygenated growth media without ceftriaxone.G93A SOD1 mouse neuroprotection: Grip strength was measured as described previously7,8. Animals were tested weekly, using a digital force meter (Columbus Instruments, Columbus, OH) for hindlimb and forelimb strength. Each grip measurement was performed three times, with a 10-20 second delay between repeats. Measurement time points began at 12 weeks of age and were repeated weekly until death (usually around 20-24 weeks of age).Survival Analysis. The primary endpoint for this preclinical trial was animal survival. Male transgenic mice expressing the human G93A SOD1 [B6SJL-TgN(SOD1-G93A)1Gur, high expressor] were bred with background-matched B6SJL wild-type females (Jackson Laboratories, Bar Harbor, ME). The progeny were genotyped and used for subsequent studies. Experiments were conducted in accordance with protocols approved by the Johns Hopkins Animal Care and Use Committee. Mice were assessed by daily observation for survival, and by weekly weighing and testing of grip strength as described above7,8. TheG93A SOD1 mice die because of respiratory failure, malnutrition and dehydration- comparable to the course of ALS in humans. To determine "survival" reliably and humanely, an artificial endpoint is used,defined by the inability of mice to right themselves 30 seconds after being placed on their sides. The moribund mice are scored as "dead", and are euthanized. The average survival for all controls (untreated G93A mice) for these experiments was 122 ± 1.9 days (sem) (n=60).Supplementary DataHippocampal glutamate trans porter currents. Glutamate transporter associated currents tended to be larger in hippocampal astrocytes following 4–7 days of treatment of postnatal rat pups with ceftriaxone (saline: 194 ± 12 pA, range, 119–292 pA, n = 14 cells; cef: 246 ± 31 pA, range, 155–574 pA, n = 15 cells; P = 0.070) although this increase did not reach stati stical significance. The lower apparent increase in transporter expression as measured through whole-cell currents, compared to biochemical methods, may reflect age-dependent changes in the properties of these juvenile astrocytes, variability in slice preparation, and/or heterogeneity in the response of individual astrocytes to ceftriaxone.Reference List1. Jackson, M. et al. Modulation of the neuronal glutamate transporter EAAT4 by twointeracting proteins. Nature410, 89-93 (2001).2. Bergles, D. E. & Jahr, C. E. Synaptic activation of glutamate transporters inhippocampal astrocytes. Neuron19, 1297-1308 (1997).3. Diamond, J. S. & Jahr, C. E. Synaptically released glutamate does not overwhelmtransporters on hippocampal astrocytes during high-frequency stimulation. J.Neurophysiol.83, 2835-2843 (2000).4. Lehre, K. P. & Danbolt, N. C. The number of glutamate transporter subtypemolecules at glutamatergic synapses: Chemical and stereological quantification in young adult rat brain. J Neurosci18, 8751-8757 (1998).5. Wadiche, J. I., Amara, S. G. & Kavanaugh, M. P. Ion fluxes associated withexcitatory amino acid transport. Neuron15, 721-728 (1995).6. Bergles, D. E., Tzingounis, A. V. & Jahr, C. E. Comparison of coupled anduncoupled currents during glutamate uptake by GLT-1 transporters. J. Neurosci.22, 10153-10162 (2002).7. Drachman, D. B. et al. Cyclooxygenase 2 inhibition protects motor neurons andprolongs survival in a transgenic mouse model of ALS. Ann. Neurol.52, 771-778 (2002).8. Kaspar, B. K., Llado, J., Sherkat, N., Rothstein, J. D. & Gage, F. H. Retrogradeviral delivery of IGF-1 prolongs survival in a mouse ALS model. Science301, 839-842 (2003).。

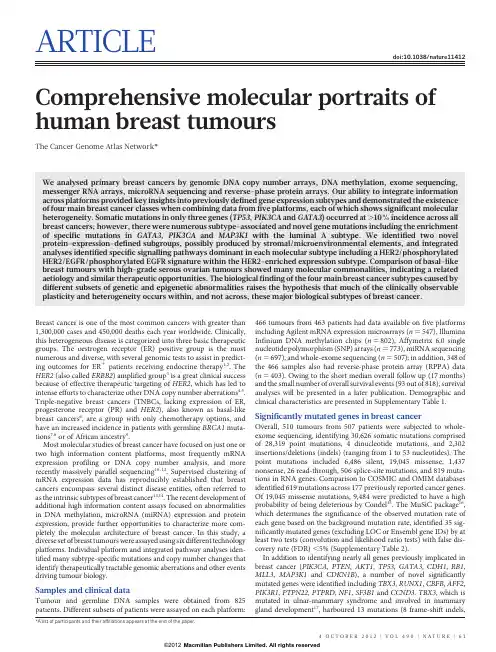
ARTICLEdoi:10.1038/nature11412 Comprehensive molecular portraits of human breast tumoursThe Cancer Genome Atlas Network*We analysed primary breast cancers by genomic DNA copy number arrays,DNA methylation,exome sequencing, messenger RNA arrays,microRNA sequencing and reverse-phase protein arrays.Our ability to integrate information across platforms provided key insights into previously defined gene expression subtypes and demonstrated the existence of four main breast cancer classes when combining data from five platforms,each of which shows significant molecular heterogeneity.Somatic mutations in only three genes(TP53,PIK3CA and GATA3)occurred at.10%incidence across all breast cancers;however,there were numerous subtype-associated and novel gene mutations including the enrichment of specific mutations in GATA3,PIK3CA and MAP3K1with the luminal A subtype.We identified two novel protein-expression-defined subgroups,possibly produced by stromal/microenvironmental elements,and integrated analyses identified specific signalling pathways dominant in each molecular subtype including a HER2/phosphorylated HER2/EGFR/phosphorylated EGFR signature within the HER2-enriched expression parison of basal-like breast tumours with high-grade serous ovarian tumours showed many molecular commonalities,indicating a related aetiology and similar therapeutic opportunities.The biological finding of the four main breast cancer subtypes caused by different subsets of genetic and epigenetic abnormalities raises the hypothesis that much of the clinically observable plasticity and heterogeneity occurs within,and not across,these major biological subtypes of breast cancer.Breast cancer is one of the most common cancers with greater than 1,300,000cases and450,000deaths each year worldwide.Clinically, this heterogeneous disease is categorized into three basic therapeutic groups.The oestrogen receptor(ER)positive group is the most numerous and diverse,with several genomic tests to assist in predict-ing outcomes for ER1patients receiving endocrine therapy1,2.The HER2(also called ERBB2)amplified group3is a great clinical success because of effective therapeutic targeting of HER2,which has led to intense efforts to characterize other DNA copy number aberrations4,5. Triple-negative breast cancers(TNBCs,lacking expression of ER, progesterone receptor(PR)and HER2),also known as basal-like breast cancers6,are a group with only chemotherapy options,and have an increased incidence in patients with germline BRCA1muta-tions7,8or of African ancestry9.Most molecular studies of breast cancer have focused on just one or two high information content platforms,most frequently mRNA expression profiling or DNA copy number analysis,and more recently massively parallel sequencing10–12.Supervised clustering of mRNA expression data has reproducibly established that breast cancers encompass several distinct disease entities,often referred to as the intrinsic subtypes of breast cancer13,14.The recent development of additional high information content assays focused on abnormalities in DNA methylation,microRNA(miRNA)expression and protein expression,provide further opportunities to characterize more com-pletely the molecular architecture of breast cancer.In this study,a diverse set of breast tumours were assayed using six different technology platforms.Individual platform and integrated pathway analyses iden-tified many subtype-specific mutations and copy number changes that identify therapeutically tractable genomic aberrations and other events driving tumour biology.Samples and clinical dataTumour and germline DNA samples were obtained from825 patients.Different subsets of patients were assayed on each platform:466tumours from463patients had data available on five platforms including Agilent mRNA expression microarrays(n5547),Illumina Infinium DNA methylation chips(n5802),Affymetrix6.0single nucleotide polymorphism(SNP)arrays(n5773),miRNA sequencing (n5697),and whole-exome sequencing(n5507);in addition,348of the466samples also had reverse-phase protein array(RPPA)data (n5403).Owing to the short median overall follow up(17months) and the small number of overall survival events(93out of818),survival analyses will be presented in a later publication.Demographic and clinical characteristics are presented in Supplementary Table1. Significantly mutated genes in breast cancerOverall,510tumours from507patients were subjected to whole-exome sequencing,identifying30,626somatic mutations comprised of28,319point mutations,4dinucleotide mutations,and2,302 insertions/deletions(indels)(ranging from1to53nucleotides).The point mutations included6,486silent,19,045missense,1,437 nonsense,26read-through,506splice-site mutations,and819muta-tions in RNA parison to COSMIC and OMIM databases identified619mutations across177previously reported cancer genes. Of19,045missense mutations,9,484were predicted to have a high probability of being deleterious by Condel15.The MuSiC package16, which determines the significance of the observed mutation rate of each gene based on the background mutation rate,identified35sig-nificantly mutated genes(excluding LOC or Ensembl gene IDs)by at least two tests(convolution and likelihood ratio tests)with false dis-covery rate(FDR),5%(Supplementary Table2).In addition to identifying nearly all genes previously implicated in breast cancer(PIK3CA,PTEN,AKT1,TP53,GATA3,CDH1,RB1, MLL3,MAP3K1and CDKN1B),a number of novel significantly mutated genes were identified including TBX3,RUNX1,CBFB,AFF2, PIK3R1,PTPN22,PTPRD,NF1,SF3B1and CCND3.TBX3,which is mutated in ulnar-mammary syndrome and involved in mammary gland development17,harboured13mutations(8frame-shift indels,*A list of participants and their affiliations appears at the end of the paper.4O C T O B E R2012|V O L490|N A T U R E|61Macmillan Publishers Limited. All rights reserved©20121in-frame deletion,1nonsense,and3missense),suggesting a loss of function.Additionally,2mutations were found in TBX4and1muta-tion in TBX5,which are genes involved in Holt–Oram syndrome18. Two other transcription factors,CTCF and FOXA1,were at or near significance harbouring13and8mutations,respectively.RUNX1and CBFB,both rearranged in acute myeloid leukaemia and interfering with haematopoietic differentiation,harboured19and9mutations, respectively.PIK3R1contained14mutations,most of which clustered in the PIK3CA interaction domain similar to previously identified mutations in glioma19and endometrial cancer20.We also observed a statistically significant exclusion pattern among PIK3R1,PIK3CA, PTEN and AKT1mutations(P50.025).Mutation of splicing factor SF3B1,previously described in myelodysplastic syndromes21and chronic lymphocytic leukaemia22,was significant with15non-silent mutations,of which4were a recurrent K700E substitution.Two protein tyrosine phosphatases(PTPN22and PTPRD)were also signifi-cantly mutated;frequent deletion/mutation of PTPRD is observed in lung adenocarcinoma23.Mutations and mRNA-expression subtype associations We analysed the somatic mutation spectrum within the context of the four mRNA-expression subtypes,excluding the normal-like group owing to small numbers(n58)(Fig.1).Several significantly mutated genes showed mRNA-subtype-specific(Supplementary Figs1–3)and clinical-subtype-specific patterns of mutation(Supplementary Table2). Significantly mutated genes were considerably more diverse and recurrent within luminal A and luminal B tumours than within basal-like and HER2-enriched(HER2E)subtypes;however,the overall mutation rate was lowest in luminal A subtype and highest in the basal-like and HER2E subtypes.The luminal A subtype harboured the most significantly mutated genes,with the most frequent being PIK3CA (45%),followed by MAP3K1,GATA3,TP53,CDH1and MAP2K4. Twelve per cent of luminal A tumours contained likely inactivating mutations in MAP3K1and MAP2K4,which represent two contiguous steps in the p38–JNK1stress kinase pathway24.Luminal B cancers exhibited a diversity of significantly mutated genes,with TP53and PIK3CA(29%each)being the most frequent.The luminal tumour subtypes markedly contrasted with basal-like cancers where TP53 mutations occurred in80%of cases and the majority of the luminal significantly mutated gene repertoire,except PIK3CA(9%),were absent or near absent.The HER2E subtype,which has frequent HER2amplification(80%),had a hybrid pattern with a high frequency of TP53(72%)and PIK3CA(39%)mutations and a much lower fre-quency of other significantly mutated genes including PIK3R1(4%). Intrinsic mRNA subtypes differed not only by mutation frequencies but also by mutation type.Most notably,TP53mutations in basal-like tumours were mostly nonsense and frame shift,whereas missense mutations predominated in luminal A and B tumours(Supplemen-tary Fig.1).Fifty-eight somatic GATA3mutations,some of which were previously described25,were detected including a hotspot2-base-pair deletion within intron4only in the luminal A subtype(13out of13 mutants)(Supplementary Fig.2).In contrast,7out of9frame-shift mutations in exon5(DNA binding domain)occurred in luminal B cancers.PIK3CA mutation frequency and spectrum also varied by mRNA subtype(Supplementary Fig.3);the recurrent PIK3CA E545K mutation was present almost exclusively within luminal A (25out of27)tumours.CDH1mutations were common(30out of 36)within the lobular histological subtype and corresponded with lower CDH1mRNA(Supplementary Fig.4)and protein expression. Finally,we identified4out of8somatic variants in HER2within lobular cancers,three of which were within the tyrosine kinase domain.We performed analyses on a selected set of genes26using the normal tissue DNA data and detected a number of germline predisposing variants.These analyses identified47out of507patients with deleterious germline variants,representing nine different genes (ATM,BRCA1,BRCA2,BRIP1,CHEK2,NBN,PTEN,RAD51C and TP53;Supplementary Table3),supporting the hypothesis that,10% of sporadic breast cancers may have a strong germlinecontribution.PIK3CA TP53MAP3K1MAP2K4GATA3MLL3CDH1PTEN PIK3R1AKT1RUNX1CBFB TBX3NCOR1CTCF FOXA1SF3B1CDKN1B RB1AFF2NF1PTPN22PTPRDCopy number statusClinical dataMutationsper MbFigure1|Significantly mutated genes and correlations with genomic andclinical features.Tumour samples are grouped by mRNA subtype:luminal A(n5225),luminal B(n5126),HER2E(n557)and basal-like(n593).Theleft panel shows non-silent somatic mutation patterns and frequencies forsignificantly mutated genes.The middle panel shows clinical features:darkgrey,positive or T2–4;white,negative or T1;light grey,N/A or equivocal.N,node status;T,tumour size.The right panel shows significantly mutatedgenes with frequent copy number amplifications(red)or deletions(blue).Thefar-right panel shows non-silent mutation rate per tumour(mutations permegabase,adjusted for coverage).The average mutation rate for eachexpression subtype is indicated.Hypermutated:mutation rates.3s.d.abovethe mean(.4.688,indicated by grey line).ARTICLE62|N A T U R E|V O L490|4O C T O B E R2012Macmillan Publishers Limited. All rights reserved©2012These data confirmed the association between the presence of germline BRCA1mutations and basal-like breast cancers7,8.Gene expression analyses(mRNA and miRNA)Several approaches were used to look for structure in the mRNA expression data.We performed an unsupervised hierarchical cluster-ing analysis of525tumours and22tumour-adjacent normal tissues using the top3,662variably expressed genes(Supplementary Fig.5); SigClust analysis identified12classes(5classes with.9samples per class).We performed a semi-supervised hierarchical cluster analysis using a previously published‘intrinsic gene list’14,which identified13 classes(9classes with.9samples per class)(Supplementary Fig.6). We also classified each sample using the50-gene PAM50model14 (Supplementary Fig.5).High concordance was observed between all three analyses;therefore,we used the PAM50-defined subtype predictor as a common classification metric.There were only eight normal-like and eight claudin-low tumours27,thus we did not per-form focussed analyses on these two subtypes.MicroRNA expression levels were assayed via Illumina sequencing, using1,222miRBase28v16mature and star strands as the reference database of miRNA transcripts/genes.Seven subtypes were identified by consensus non-negative matrix factorization(NMF)clustering using an abundance matrix containing the25%most variable miRNAs(306transcripts/genes or MIMATs(miRNA IDs)).These subtypes correlated with mRNA subtypes,ER,PR and HER2clinical status(Supplementary Fig.7).Of note,miRNA groups4and5 showed high overlap with the basal-like mRNA subtype and con-tained many TP53mutations.The remaining miRNA groups(1–3, 6and7)were composed of a mixture of luminal A,luminal B and HER2E with little correlation with the PAM50defined subtypes.With the exception of TP53—which showed a strong positive correlation—and PIK3CA and GATA3—which showed negative associations with groups4and5,respectively—there was little correlation with muta-tion status and miRNA subtype.DNA methylationIllumina Infinium DNA methylation arrays were used to assay802 breast tumours.Data from HumanMethylation27(HM27)and HumanMethylation450(HM450)arrays were combined and filtered to yield a common set of574probes used in an unsupervised clustering analysis,which identified five distinct DNA methylation groups (Supplementary Fig.8).Group3showed a hypermethylated pheno-type and was significantly enriched for luminal B mRNA subtype and under-represented for PIK3CA,MAP3K1and MAP2K4mutations. Group5showed the lowest levels of DNA methylation,overlapped with the basal-like mRNA subtype,and showed a high frequency of TP53mutations.HER2-positive(HER21)clinical status,or the HER2E mRNA subtype,had only a modest association with the methylation subtypes.A supervised analysis of the DNA methylation and mRNA expres-sion data was performed to compare DNA methylation group3 (N549)versus all tumours in groups1,2and4(excluding group5, which consisted predominantly of basal-like tumours).This analysis identified4,283genes differentially methylated(3,735higher in group 3tumours)and1,899genes differentially expressed(1,232downregu-lated);490genes were both methylated and showed lower expression in group3tumours(Supplementary Table4).A DAVID(database for annotation,visualization and integrated discovery)functional annota-tion analysis identified‘extracellular region part’and‘Wnt signalling pathway’to be associated with this490-gene set;the group3hyper-methylated samples showed fewer PIK3CA and MAP3K1mutations, and lower expression of Wnt-pathway genes.DNA copy numberA total of773breast tumours were assayed using Affymetrix6.0 SNP arrays.Segmentation analysis and GISTIC were used to identify focal amplifications/deletions and arm-level gains and losses (Supplementary Table5).These analyses confirmed all previously reported copy number variations and highlighted a number of sig-nificantly mutated genes including focal amplification of regions con-taining PIK3CA,EGFR,FOXA1and HER2,as well as focal deletions of regions containing MLL3,PTEN,RB1and MAP2K4(Supplementary Fig.9);in all cases,multiple genes were included within each altered region.Importantly,many of these copy number changes correlated with mRNA subtype including characteristic loss of5q and gain of 10p in basal-like cancers5,29and gain of1q and/or16q loss in luminal tumours4.NMF clustering of GISTIC segments identified five copy number clusters/groups that correlated with mRNA subtypes,ER,PR and HER2clinical status,and TP53mutation status(Supplementary Fig.10).In addition,this aCGH subtype classification was highly correlated with the aCGH subtypes recently defined by ref.30 (Supplementary Fig.11).Reverse phase protein arraysQuantified expression of171cancer-related proteins and phospho-proteins by RPPA was performed on403breast tumours31. Unsupervised hierarchical clustering analyses identified seven subtypes;one class contained too few cases for further analysis (Supplementary Fig.12).These protein subtypes were highly concordant with the mRNA subtypes,particularly with basal-like and HER2E mRNA subtypes.Closer examination of the HER2-containing RPPA-defined subgroup showed coordinated overexpres-sion of HER2and EGFR with a strong concordance with phosphorylated HER2(pY1248)and EGFR(pY992),probably from heterodimeriza-tion and cross-phosphorylation.Although there is a potential for modest cross reactivity of antibodies against these related total and phospho-proteins,the concordance of phosphorylation of HER2and EGFR was confirmed using multiple independent antibodies.In RPPA-defined luminal tumours,there was high protein expres-sion of ER,PR,AR,BCL2,GATA3and INPP4B,defining mostly luminal A cancers and a second more heterogeneous protein subgroup composed of both luminal A and luminal B cancers.Two potentially novel protein-defined subgroups were identified:reactive I consisted primarily of a subset of luminal A tumours,whereas reactive II consisted of a mixture of mRNA subtypes.These groups are termed ‘reactive’because many of the characteristic proteins are probably produced by the microenvironment and/or cancer-activated fibroblasts including fibronectin,caveolin1and collagen VI.These two RPPA groups did not have a marked difference in the percentage tumour cell content when compared to each other,or the other protein subtypes, as assessed by SNP array analysis or pathological examination.In addition,supervised analyses of reactive I versus II groups using miRNA expression,DNA methylation,mutation,or DNA copy number data identified no significant differences between these groups, whereas similar supervised analyses using protein and mRNA expres-sion identified many differences.Multiplatform subtype discoveryTo reveal higher-order structure in breast tumours based on multiple data types,significant clusters/subtypes from each of five platforms were analysed using a multiplatform data matrix subjected to unsupervised consensus clustering(Fig.2).This‘cluster of clusters’(C-of-C)approach illustrated that basal-like cancers had the most distinct multiplatform signature as all the different platforms for the basal-like groups clustered together.To a great extent,the four major C-of-C subdivisions correlated well with the previously published mRNA subtypes(driven,in part,by the fact that the four intrinsic subtypes were one of the inputs).Therefore,we also performed C-of-C analysis with no mRNA data present(Supplementary Fig.13) or with the12unsupervised mRNA subtypes(Supplementary Fig.14), and in each case4–6groups were identified.Recent work identified ten copy-number-based subgroups in a997breast cancer set30.We evaluatedARTICLE4O C T O B E R2012|V O L490|N A T U R E|63Macmillan Publishers Limited. All rights reserved ©2012this classification in a C-of-C analysis instead of our five-class copy number subtypes,with either the PAM50(Supplementary Fig.15)or 12unsupervised mRNA subtypes (Supplementary Fig.16);each of these C-of-C classifications was highly correlated with PAM50mRNA subtypes and with the other C-of-C analyses (Fig.2).The transcriptional profiling and RPPA platforms demonstrated a high correlation with the consensus structure,indicating that the informa-tion content from copy number aberrations,miRNAs and methylation is captured at the level of gene expression and protein function.Luminal/ER 1summary analysisLuminal/ER 1breast cancers are the most heterogeneous in terms of gene expression (Supplementary Fig.5),mutation spectrum (Fig.1),copy number changes (Supplementary Fig.9)and patient outcomes 1,14.One of the most dominant features is high mRNA and protein expres-sion of the luminal expression signature (Supplementary Fig.5),which contains ESR1,GATA3,FOXA1,XBP1and MYB ;the luminal/ER 1cluster also contained the largest number of significantly mutated genes.Most notably,GATA3and FOXA1were mutated in a mutually exclusive fashion,whereas ESR1and XBP1were typically highly expressed but infrequently mutated.Mutations in RUNX1and its dimerization partner CBFB may also have a role in aberrant ER signalling in luminal tumours,as RUNX1functions as an ER ‘DNA tethering factor’32.PARADIGM 33analysis comparing luminal versus basal-like cancers further emphasized the presence of a hyperactivated FOXA1–ER complex as a critical network hub differentiating these two tumour subtypes (Supplementary Fig.17).A confirmatory finding here was the high mutation frequency of PIK3CA in luminal/ER 1breast cancers 34,35.Through multipletechnology platforms,we examined possible relationships between PIK3CA mutation,PTEN loss,INPP4B loss and multiple gene and protein expression signatures of pathway activity.RPPA data demonstrated that pAKT,pS6and p4EBP1,typical markers of phosphatidylinositol-3-OH kinase (PI(3)K)pathway activation,were not elevated in PIK3CA -mutated luminal A cancers;instead,they were highly expressed in basal-like and HER2E mRNA subtypes (the latter having frequent PIK3CA mutations)and correlated strongly with INPP4B and PTEN loss,and to a degree with PIK3CA amplification.Similarly,protein 36and three mRNA signatures 37–39of PI(3)K pathway activation were enriched in basal-like over luminal A cancers (Fig.3a).This apparent disconnect between the presence of PIK3CA mutations and biomarkers of pathway activation has been previously noted 36.Another striking luminal/ER 1subtype finding was the frequent mutation of MAP3K1and MAP2K4,which represent two contiguous steps within the p38–JNK1pathway 24,40.These mutations are predicted to be inactivating,with MAP2K4also a target of focal DNA loss in luminal tumours (Supplementary Fig.9).To explore the possible interplay between PIK3CA ,MAP3K and MAP2K4signalling,MEMo analysis 41was performed to identify mutually exclusive alterations targeting frequently altered genes likely to belong to the same pathway (Fig.4).Across all breast cancers,MEMo identified a set of modules that highlight the differential activation events within the receptor tyrosine kinase (RTK)–PI(3)K pathway (Fig.4a);mutations of PIK3CA were very common in luminal/ER 1cancers whereas PTEN loss was more common in basal-like tumours.Almost all MAP3K1and MAP2K4mutations were in luminal tumours,yet MAP3K1and MAP2K4appeared almost mutually exclusive relative to one another.The TP53pathway was differentially inactivated in luminal/ER 1breast cancers,with a low TP53mutation frequency in luminal A (12%)and a higher frequency in luminal B (29%)cancers (Fig.1).In addition to TP53itself,a number of other pathway-inactivating events occurred including ATM loss and MDM2amplification (Figs 3b and 4b),both of which occurred more frequently within luminal B cancers.Gene expression analysis demonstrated that individual markers of functional TP53(GADD45A and CDKN1A ),and TP53activity 42,43signatures,were highest in luminal A cancers (Fig.3b).These data indicate that the TP53pathway remains largely intact in luminal A cancers but is often inactivated in the more aggressive luminal B cancers 44.Other PARADIGM-based pathway differences driving luminal B versus luminal A included hyperactivation of transcriptional activity associated with MYC and FOXM1proliferation.The critical retinoblastoma/RB1pathway also showed mRNA-subtype-specific alterations (Fig.3c).RB1itself,by mRNA and protein expression,was detectable in most luminal cancers,with highest levels within luminal A.A common oncogenic event was cyclin D1amplification and high expression,which preferentially occurred within luminal tumours,and more specifically within luminal B.In contrast,the presumed tumour suppressor CDKN2C (also called p18)was at its lowest levels in luminal A cancers,con-sistent with observations in mouse models 45.Finally,RB1activity signatures were also high in luminal cancers 46–48.Luminal A tumours,which have the best prognosis,are the most likely to retain activity of the major tumour suppressors RB1and TP53.These genomic characterizations also provided clues for druggable targets.We compiled a drug target table in which we defined a target as a gene/protein for which there is an approved or investigational drug in human clinical trials targeting the molecule or canonical pathway (Supplementary Table 6).In luminal/ER 1cancers,the high frequency of PIK3CA mutations suggests that inhibitors of this activated kinase or its signalling pathway may be beneficial.Other potential significantly mutated gene drug candidates include AKT1inhibitors (11out of 12AKT1variants were luminal)and PARP inhibitors for BRCA1/BRCA2mutations.Although still unapproved as biomarkers,many potential copy-number-based drugtargetsP <0.001P <0.001P <0.001P <0.001P =0.002P =0.01P <0.001 P <0.001 P <0.001 P <0.001 P =0.02miRNA 2Methy 2CN 2PAM50 LumA RPPA LumA Methy 1miRNA 6RPPA reactive I CN 4RPPA reactive II miRNA 3CN 1Methy 5PAM50 basal RPPA basal miRNA 5PAM50 normal CN 3Methy 4miRNA 4PAM50 HER2RPPA HER2CN 5Methy 3PAM50 LumB RPPA LumA/B miRNA 7miRNA 1acbFigure 2|Coordinated analysis of breast cancer subtypes defined from five different genomic/proteomic platforms.a ,Consensus clustering analysis of the subtypes identifies four major groups (samples,n 5348).The blue and white heat map displays sample consensus.b ,Heat-map display of the subtypes defined independently by miRNAs,DNA methylation,copy number (CN),PAM50mRNA expression,and RPPA expression.The red bar indicates membership of a cluster type.c ,Associations with molecular and clinical features.P values were calculated using a chi-squared test.ARTICLE64|N A T U R E |V O L 490|4O C T O B E R 2012Macmillan Publishers Limited. All rights reserved©2012were identified including amplifications of fibroblast growth factor receptors (FGFRs)and IGFR1,as well as cyclin D1,CDK4and CDK6.A summary of the general findings in luminal tumours and the other subtypes is presented in Table 1.HER2-based classifications and summary analysisDNA amplification of HER2was readily evident in this study (Supplementary Fig.9)together with overexpression of multipleHER2-amplicon-associated genes that in part define the HER2E mRNA subtype (Supplementary Fig.5).However,not all clinically HER21tumours are of the HER2E mRNA subtype,and not all tumours in the HER2E mRNA subtype are clinically HER21.Integrated analysis of the RPPA and mRNA data clearly identified a HER21group (Supplementary Fig.12).When the HER21protein and HER2E mRNA subtypes overlapped,a strong signal of EGFR,pEGFR,HER2and pHER2was observed.However,only ,50%of clinically HER21tumours fall into this HER2E-mRNA-subtype/HER2-protein group,the rest of the clinically HER21tumours were observed predominantly in the luminal mRNA subtypes.These data indicate that there exist at least two types of clinically defined HER21tumours.To identify differences between these groups,a supervised gene expression analysis comparing 36HER2E-mRNA-subtype/HER21versus 31luminal-mRNA-subtype/HER21tumours was performed and identified 302differentially expressed genes (q -value 50%)(Supplementary Fig.18and Supplementary Table 7).These genes largely track with ER status but also indicated that HER2E-mRNA-subtype/HER21tumours showed significantly higher expres-sion of a number of RTKs including FGFR4,EGFR ,HER2itself,as well as genes within the HER2amplicon (including GRB7).Conversely,the luminal-mRNA-subtype/HER21tumours showed higher expression of the luminal cluster of genes including GATA3,BCL2and ESR1.Further support for two types of clinically defined HER21disease was evident in the somatic mutation data supervised by either mRNA subtype or ER status;TP53mutations were significantly enriched in HER2E or ER-negative tumours whereas GATA3muta-tions were only observed in luminal subtypes or ER 1tumours.Analysis of the RPPA data according to mRNA subtype identified 36differentially expressed proteins (q -value ,5%)(Supplementary Fig.18G and Supplementary Table 8).The EGFR/pEGFR/HER2/pHER2signal was again observed and present within the HER2E-mRNA-subtype/HER21tumours,as was high pSRC and pS6;con-versely,many protein markers of luminal cancers again distinguished the luminal-mRNA-subtype/HER21tumours.Given the importance of clinical HER2status,a more focused analysis was performed based on the RPPA-defined protein expression of HER2(Supplementary Fig.19)—the results strongly recapitulated findings from the RPPA and mRNA subtypes including a high correlation between HER2clinical status,HER2protein by RPPA,pHER2,EGFR and pEGFR.These multiple signatures,namely HER2E mRNA subtype,HER2amplicon genes by mRNA expression,and RPPA EGFR/pEGFR/HER2/pHER2signature,ultimately identify at least two groups/subtypes within clinically HER21tumours (Table 1).These signatures represent breast cancer biomarker(s)that could potentially predict response to anti-HER2targeted therapies.Many therapeutic advances have been made for clinically HER21disease.This study has identified additional somatic mutations that represent potential therapeutic targets within this group,including a high frequency of PIK3CA mutations (39%),a lower frequency of PTEN and PIK3R1mutations (Supplementary Table 6),and genomic losses of PTEN and INPP4B .Other possible druggable mutations included variants within HER family members including two somatic mutations in HER2,two within EGFR ,and five within HER3.Pertuzumab,in combination with trastuzumab,targets the HER2–HER3heterodimer 49;however,these data suggest that targeting EGFR with HER2could also be beneficial.Finally,the HER2E mRNA subtype typically showed high aneuploidy,the highest somatic mutation rate (Table 1),and DNA amplification of other potential therapeutic targets including FGFRs,EGFR ,CDK4and cyclin D1.Basal-like summary analysisThe basal-like subtype was discovered more than a decade ago by first-generation cDNA microarrays 13.These tumours are often referred to as triple-negative breast cancers (TNBCs)because most basal-like tumours are typically negative for ER,PR andHER2.IARC GSK KANNAN TROESTERCDKN1A mRNA GADD45A mRNAMDM2 mRNATP53 copy CDKN2A copy CHEK1 copy ATM copy MDM2 copy MDM4 copy PPM1D copy TP53 mutationMissense Nonsense/frame shift Splice siteTP53 LOH Differences by TP53 mutation (P <0.0001)Differences by luminal A subtype vs others (P <0.0001)TP53 pathway (506 tumours with mRNA/mutation data)b Gain Loss Copy change Gene signature activity More Less mRNA expression Low High High Low Protein expression mRNA subtype:Basal-likeLuminal BLuminal ANormal-likePIK3CA mutation PTEN LOH INPP4B LOH Correlated with PI(3)K protein signature (P <0.0005)Differences by PTEN/INPP4B LOH (P <0.05)Differences by basal subtype vs others (P <0.01)pAkt 308pAkt 473pmTOR pGSK3p70S6Kp389pS6 240.244p4EBP1 65INPP4B PTENRPPA (protein)Saal (mRNA)CMap (mRNA)Majumder (mRNA)PI(3)K pathway (390 tumours with mRNA/mutation/protein data)a PTEN copy INPP4B copy PIK3CA copy CHICAS LARAHERSCHKOWITZRB1 mRNA CDKN2A mRNA CDKN2C mRNARB1 copy CCND1 copy CCNE1 copy RB1 mutationRB1 LOH Differences by RB1 mutation (P <0.003)Differences by RB1 LOH (P <0.005)RB1 protein Differences by luminal A subtype vs others (P <0.0001)RB pathway (506 tumours with mRNA/mutation data)c HER2-enriched*‡* † ‡* † ‡* † ‡* † ‡† ‡† ‡† ‡††† ‡† ‡† ‡† ‡* † ‡* † ‡*‡†† ‡† ‡* † ‡† ‡‡* † ‡* † ‡* † ‡* † ‡* ††* †* †* †* †* †*†*†*†‡*†‡Figure 3|Integrated analysis of the PI(3)K,TP53and RB1pathways.Breast cancer subtypes differ by genetic and genomic targeting events,withcorresponding effects on pathway activity.a –c ,For PI(3)K (a ),TP53(b )and RB1(c )pathways,key genes were selected using prior biological knowledge.Multiple mRNA expression signatures for a given pathway were defined (details in Supplementary Methods;PI(3)K:Saal,PTEN loss in human breast tumours;CMap,PI(3)K/mTOR inhibitor treatment in vitro ;Majumder,Akt overexpression in mouse model;TP53:IARC,expert-curated p53targets;GSK,TP53mutant versus wild-type cell lines;KANNAN,TP53overexpression in vitro ;TROESTER,TP53knockdown in vitro ;RB:CHICAS,RB1mouse knockout versus wild type;LARA,RB1knockdown in vitro ;HERSCHKOWITZ,RB1loss of heterozygosity (LOH)in human breast tumours)and applied to the gene expression data,in order to score each tumour for relative signature activity (yellow,more active).The PI(3)K panel includes a protein-based (RPPA)proteomic signature.Tumours were ordered first by mRNA subtype,although specific ordering differs between the panels.P values were calculated by a Pearson’s correlation or a Chi-squared test.ARTICLE 4O C T O B E R 2012|V O L 490|N A T U R E |65Macmillan Publishers Limited. All rights reserved©2012。

!N"!急性胰腺炎胰酶异常活化和分泌的分子机制崔恬玉a,刘瑞霞b,阴 宏a首都医科大学附属北京妇产医院a.内科,b.中心试验室,北京100026摘要:胰腺腺泡细胞内胰酶异常活化和分泌是急性胰腺炎(AP)的重要发病机制之一,可直接损伤胰腺组织加速疾病进程,诱发重症急性胰腺炎。
目前临床抑制胰酶异常活化和分泌的药物效果欠佳,探寻新的治疗靶点十分重要。
本文归纳了AP胰酶异常活化和分泌的病理事件(胞浆钙离子超载、溶酶体与酶原颗粒的共定位、细胞器损伤、胰酶顶端侧分泌受阻和基底侧分泌增加等),搜集了相关事件的分子机制,讨论了胰酶异常活化和分泌在AP早期的作用过程,为未来靶向药物的研发提供思路。
关键词:胰腺炎;胰酶;分泌小泡基金项目:北京市教育委员会科技计划一般项目(KM201910025007);国家自然科学基金(81571933)ResearchadvancesinabnormalactivationandsecretionofpancreaticenzymesinacutepancreatitisCUITianyua,LIURuixiab,YINChenghonga.(a.DepartmentofInternalMedicine,b.DepartmentofCentralLaboratory,BeijingObstetricsandGynecologyHospital,CapitalMedicalUniversity,Beijing100026,China)Correspondingauthor:YINChenghong,yinchh@ccmu.edu.cn(ORCID:0000-0002-2503-3285)Abstract:Abnormalactivationandsecretionofpancreaticenzymesinpancreaticacinarcellsisoneoftheimportantpathogenesesofacutepancreatitis(AP)andcandirectlydamagethepancreatictissuetoacceleratediseaseprogressionandinducesevereAP.Atpresent,thedrugsinhibitingtheabnormalactivationandsecretionofpancreaticenzymestendtohaveanunsatisfactoryeffectinclinicalpractice,andtherefore,itisofgreatimportancetosearchfornewtherapeutictargets.Thisarticlesummarizesthepathologicaleventsofabnormalactiva tionandsecretionofpancreaticenzymes(cytoplasmiccalciumoverload,colocalizationoflysosomesandzymogengranules,organelleinjury,obstructedapicalsecretionoftrypsin,andincreasedbasalsecretionoftrypsin),collectsthemolecularmechanismsofrelatedevents,anddiscussestheroleofabnormalactivationandsecretionofpancreaticenzymesintheearlystageofAP,soastoprovideideasforthedevelopmentoftargeteddrugsinthefuture.Keywords:Pancreatitis;Pancreatin;SecretoryVesiclesResearchfunding:BeijingMunicipalCommissionofEducation(KM201910025007);NationalNaturalScienceFoundationofChina(81571933)DOI:10.3969/j.issn.1001-5256.2022.05.047收稿日期:2021-08-31;录用日期:2021-10-08通信作者:阴宏,yinchh@ccmu.edu.cn 急性胰腺炎(acutepancreatitis,AP)是一种临床常见急腹症,发病率为34/10万,在世界范围内呈上升趋势[1],其中重症急性胰腺炎(severeacutepancreatitis,SAP)常合并严重并发症,病死率达15%~35%[2-3],严重威胁人民生命健康,并带来巨大经济负担。

!K"!Hippo/YAP参与肝纤维化发生发展的作用机制赵晓璐,张春艳,高晓阳,马月宏内蒙古医科大学基础医学院,呼和浩特010110摘要:多种病因引起肝脏微环境的破坏而致肝组织结构功能丧失,启动肝纤维化进程,主要以肝星状细胞(HSC)活化为肌成纤维细胞,分泌大量以胶原为主的细胞外基质(ECM)为特征表现。
虽然目前对抗肝纤维化机制的研究已有很多,但仍缺乏有效的靶点药物应用于临床,近年来关于抗肝纤维化的研究多集中在肝纤维化进展期的干预,而忽略了早期肝纤维化的具体机制如何。
最近关于Hippo信号在肝纤维化中的研究逐渐增多,集中于核心转录因子Yes相关蛋白(YAP)在早期活化HSC中的表达,及该通路调控HSC的状态,本文主要介绍了Hippo/YAP通路参与调控肝纤维化早期及进展期的作用,简述了调控核心因子YAP的稳定表达与核转位可逆转肝纤维化的潜在作用,表明了该通路可为临床治疗提供新的方向及靶点。
关键词:肝硬化;Hippo通路;Yes相关蛋白;病理过程基金项目:国家自然科学基金(81960759,81560706);内蒙古自治区自然科学基金资助项目(2019MS08010,2014MS0841);内蒙古自治区草原英才培养计划;内蒙古自治区教坛新秀培养计划;内蒙古医科大学致远人才计划项目MechanismofactionoftheHippo/YAPpathwayinthedevelopmentandprogressionofliverfibrosisZHAOXiaolu,ZHANGChunyan,GAOXiaoyang,MAYuehong.(InnerMongoliaMedicalUniversitySchoolofBasicMedicine,Hohhot010110,China)Correspondingauthor:MAYuehong,myh19982002@sina.com(ORCID:0000-0003-1414-0066)Abstract:Variousetiologiescausethedestructionoflivermicroenvironment,whichleadstothelossofliverstructureandfunctionandinitiatestheprocessofliverfibrosis.Hepaticstellatecells(HSCs)aremainlyactivatedintomyofibroblaststhatsecretealargeamountofextracellularmatrix(ECM),mostlycollagen.Althoughtherehavebeenmanystudiesontheanti-liverfibrosismechanism,thereisstillalackofeffectivetargetdrugsforclinicalapplication,andinrecentyears,studiesonanti-liverfibrosishavemainlyfocusedoninterventionsfortheadvancedstageofliverfibrosis,whileignoringthespecificmechanismofearly-stageliverfibrosis.Recently,therehavebeenanin creasingnumberofstudiesonHipposignalinginliverfibrosis,withafocusontheexpressionofthecoretranscriptionfactorYes-associatedprotein(YAP)inearlyactivatedHSCsandtheregulationofHSCstatusbythispathway.ThisarticlemainlyintroducestheroleoftheHip po/YAPpathwayintheregulationofearly-stageoradvancedliverfibrosisandbrieflydescribesthepotentialroleofregulatingthestableexpressionandnucleartranslocationofthecoretranscriptionfactorYAPinreversingliverfibrosis,suggestingthatthispathwaycanprovidenewdirectionsandtargetsforclinicaltreatment.Keywords:LiverCirrhosis;HippoPathway;Yes-associatedProtein;PathologicProcessesResearchfunding:NationalNaturalScienceFoundationofChina(81960759,81560706);NaturalScienceFoundationofInnerMongoliaAutonomousRegion(2019MS08010,2014MS0841);InnerMongoliaAutonomousRegionGrasslandTalentsTrainingProgram;InnerMon goliaAutonomousRegionTeachingRookieTrainingProgram;InnerMongoliaMedicalUniversityZhiyuanTalentProgramDOI:10.3969/j.issn.1001-5256.2022.07.037收稿日期:2021-12-13;录用日期:2022-01-13通信作者:马月宏,myh19982002@sina.com 多种外界因素刺激而致的肝纤维化均伴随着成纤维细胞激活及细胞外基质(ECM)增加,同时引起多种细胞因子、趋化因子的释放,进一步促进肝星状细胞(HSC)活化,导致异常沉积在细胞表面的基质硬度增加形成的正反馈回路[1],放大了肝纤维化的过程。

expressionofinterferonregulatoryfactor9(IRF9)proteinaswellasmyxovirusresistanceprotein1(MxA)proteinintheJAK/STATsignalingpathway.Results InHepG2cellsexpressingHBVtransientlyandHepG2 2 15cellsstablyexpressingHBV,theexpressionofPTENproteinbothdecreased;theexpressionofHBV relatedantigensandHBVpgRNAdecreasedinPTEN OEHepG2 2 15cellscomparedwiththecontrolgroup.Afterthetreatmentbypoly(I∶C),thelevelofIFN αmRNAwassignificantlyhigherthanthatofthecontrolgroup,andtheexpressionofIRF9andMxAptoteinrelatedtotheJAK/STATsignalingpathwaybothincreased.Conclusion HBVmayplayaroleinantagonizingtheantiviralactivityofIFN αbydown regulatingtheexpressionofPTEN.Keywords hepatitisBvirus;PTEN;IFN α;JAK/STATsignalingpathway;chronichepatitisB网络出版时间:2022-05-2814:26 网络出版地址:https://kns.cnki.net/kcms/detail/34.1065.R.20220526.1018.021.html达格列净通过丝氨酸/苏氨酸蛋白激酶/内皮型一氧化氮合酶途径调节高糖环境下的内皮祖细胞功能解丹丹,巫婷婷,赵晓彤,许慕蓉,陈明卫摘要 目的 探讨达格列净(DAPA)对高糖环境下体外培养的大鼠内皮祖细胞(EPCs)功能的影响。

Sample Selection Bias as a Specification ErrorJames J.HeckmanEconometrica,Vol.47,No.1.(Jan.,1979),pp.153-161.Stable URL:/sici?sici=0012-9682%28197901%2947%3A1%3C153%3ASSBAAS%3E2.0.CO%3B2-J Econometrica is currently published by The Econometric Society.Your use of the JSTOR archive indicates your acceptance of JSTOR's Terms and Conditions of Use,available at/about/terms.html.JSTOR's Terms and Conditions of Use provides,in part,that unless you have obtained prior permission,you may not download an entire issue of a journal or multiple copies of articles,and you may use content in the JSTOR archive only for your personal,non-commercial use.Please contact the publisher regarding any further use of this work.Publisher contact information may be obtained at/journals/econosoc.html.Each copy of any part of a JSTOR transmission must contain the same copyright notice that appears on the screen or printed page of such transmission.JSTOR is an independent not-for-profit organization dedicated to and preserving a digital archive of scholarly journals.For more information regarding JSTOR,please contact support@.Wed Apr1811:20:522007You have printed the following article:Sample Selection Bias as a Specification ErrorJames J.HeckmanEconometrica ,Vol.47,No.1.(Jan.,1979),pp.153-161.Stable URL:/sici?sici=0012-9682%28197901%2947%3A1%3C153%3ASSBAAS%3E2.0.CO%3B2-JThis article references the following linked citations.If you are trying to access articles from anoff-campus location,you may be required to first logon via your library web site to access JSTOR.Please visit your library's website or contact a librarian to learn about options for remote access to JSTOR.[Footnotes]2Wage Comparisons--A Selectivity BiasReuben GronauThe Journal of Political Economy ,Vol.82,No.6.(Nov.-Dec.,1974),pp.1119-1143.Stable URL:/sici?sici=0022-3808%28197411%2F12%2982%3A6%3C1119%3AWCSB%3E2.0.CO%3B2-L2Comments on Selectivity Biases in Wage ComparisonsH.Gregg LewisThe Journal of Political Economy ,Vol.82,No.6.(Nov.-Dec.,1974),pp.1145-1155.Stable URL:/sici?sici=0022-3808%28197411%2F12%2982%3A6%3C1145%3ACOSBIW%3E2.0.CO%3B2-YReferences1Regression Analysis when the Dependent Variable Is Truncated NormalTakeshi AmemiyaEconometrica ,Vol.41,No.6.(Nov.,1973),pp.997-1016.Stable URL:/sici?sici=0012-9682%28197311%2941%3A6%3C997%3ARAWTDV%3E2.0.CO%3B2-FLINKED CITATIONS -Page 1of 2-2Specification Bias in Estimates of Production FunctionsZvi GrilichesJournal of Farm Economics ,Vol.39,No.1.(Feb.,1957),pp.8-20.Stable URL:/sici?sici=1071-1031%28195702%2939%3A1%3C8%3ASBIEOP%3E2.0.CO%3B2-V 4Wage Comparisons--A Selectivity BiasReuben GronauThe Journal of Political Economy ,Vol.82,No.6.(Nov.-Dec.,1974),pp.1119-1143.Stable URL:/sici?sici=0022-3808%28197411%2F12%2982%3A6%3C1119%3AWCSB%3E2.0.CO%3B2-L8Dummy Endogenous Variables in a Simultaneous Equation SystemJames J.HeckmanEconometrica ,Vol.46,No.4.(Jul.,1978),pp.931-959.Stable URL:/sici?sici=0012-9682%28197807%2946%3A4%3C931%3ADEVIAS%3E2.0.CO%3B2-F9Asymptotic Properties of Non-Linear Least Squares EstimatorsRobert I.JennrichThe Annals of Mathematical Statistics ,Vol.40,No.2.(Apr.,1969),pp.633-643.Stable URL:/sici?sici=0003-4851%28196904%2940%3A2%3C633%3AAPONLS%3E2.0.CO%3B2-311Comments on Selectivity Biases in Wage ComparisonsH.Gregg LewisThe Journal of Political Economy ,Vol.82,No.6.(Nov.-Dec.,1974),pp.1145-1155.Stable URL:/sici?sici=0022-3808%28197411%2F12%2982%3A6%3C1145%3ACOSBIW%3E2.0.CO%3B2-Y 12Specification Errors and the Estimation of Economic RelationshipsH.TheilRevue de l'Institut International de Statistique /Review of the International Statistical Institute ,Vol.25,No.1/3.(1957),pp.41-51.Stable URL:/sici?sici=0373-1138%281957%2925%3A1%2F3%3C41%3ASEATEO%3E2.0.CO%3B2-BLINKED CITATIONS -Page 2of 2-。

S1PR1-mediated IFNAR1 degradation modulates plasmacytoiddendritic cell interferon-α autoamplification由S1PR1介导的IFNAR1降解可以调节浆细胞样树突状细胞α-干扰素的自动扩增/信号放大摘要:Blunting immunopathology without abolishing host defense is the foundation for safe and effective modulation of infectious and autoimmune diseases.没有废除宿主防御机制的免疫病理钝化是安全、有效调节传染病和自身免疫性疾病的基础。
Sphingosine 1-phosphate receptor 1 (S1PR1) agonists are effective in treating infectious and multiple autoimmune pathologies; however, mechanisms underlying their clinical efficacy are yet to be fully elucidated.1-磷酸-鞘氨醇受体1(S1PR1)促效药对于治疗传染病和多种自身免疫性疾病是有效的,然而,其临床疗效的具体机制尚未被完全阐明。
Here, we uncover an unexpected mechanism of convergence between S1PR1 and interferon alpha receptor 1 (IFNAR1) signaling pathways.在本研究中,我们意外发现S1PR1与α-干扰素受体1(IFNAR1)信号通路之间的趋同/聚集机制。
Activation of S1PR1 signaling by pharmacological tools or endogenous ligand sphingosine-1 phosphate (S1P) inhibits type 1 IFN responses that exacerbate numerous pathogenic conditions.通过药理作用或内源性配体1-磷酸-鞘氨醇(S1P)发出信号激活S1PR1可以抑制1型干扰素应答,这将提供大量致病条件。
Incorporationofamatrixmetalloproteinase-sensitivesubstrateintoself-assemblingpeptideseAmodelforbiofunctionalscaffolds
YingChaua,YingLuoh,AlexC.Y.Cheunga,YusukeNagaic,ShuguangZhangd,e,JamesB.Koblerf,g,StevenM.Zeitelsf,g,RobertLangerb,*
aDepartmentofChemicalEngineering,TheHongKongUniversityofScienceandTechnology,ClearWaterBay,HongKong,China
bDepartmentofChemicalEngineering,MassachusettsInstituteofTechnology,77MassachusettsAvenue,E25-342,Cambridge,MA02139,USA
cMeniconCo.,Ltd.,5-1-10Takamoridai,Kasugai,Aichi487-0032,Japan
dCenterforBiomedicalEngineering,MassachusettsInstituteofTechnology,77MassachusettsAvenue,NE47-379,Cambridge,MA02139,USA
eCenterforBitsandAtoms,MassachusettsInstituteofTechnology,77MassachusettsAvenue,Cambridge,MA02139,USA
fDepartmentofSurgery,HarvardMedicalSchool,Boston,MA,USA
gCenterforLaryngealSurgeryandVoiceRehabilitation,MassachusettsGeneralHospital,OneBowdoinSquare,11thfloor,Boston,MA02114,USA
hDepartmentofBiomedicalEngineering,PekingUniversity,60Yan-NanYuan,HaiDianDistrict,Beijing,China
Received29May2007;accepted6November2007Availableonline14January2008
AbstractControllingandguidingcellbehaviorrequiresscaffoldingmaterialscapableofprogrammingthethree-dimensional(3-D)extracellularenvi-ronment.Inthisstudy,wedevisedanewself-assemblingpeptidetemplateforsynthesizingnanofibroushydrogelscontainingcell-responsiveli-gands.Inparticular,theinsertionofamatrixmetalloproteinase-2(MMP-2)labilehexapeptideintotheself-assemblingbuildingblocksofarginine-alanine-aspartate-alanine(RADA)wasinvestigated.Aseriesofpeptides,variedbythepositionoftheMMP-2hexapeptidesubstrateandthelengthofRADAblocks,werepreparedbyparallelsynthesis.Theirself-assemblingcapabilitieswerecharacterizedandcomparedbycir-culardichroismspectroscopyanddynamicalmechanicalanalysis.Amongallthedifferentinsertionpatterns,thesequencecomprisingacentricallypositionedMMP-2substratewasflankedwiththreeRADAunitsoneachsideself-assembledintoahydrogelmatrix,withmechanicalpropertiesandnanofibermorphologycomparabletothenativematerialbuiltwith(RADA)4alone.ExposureofthenewgeltoMMP-2resultedinpeptide
cleavage,asconfirmedbymassspectroscopy,andadecreaseinsurfacehardness,asdetectedbynanoindentor,indicatingthattheenzymeme-diateddegradationwaslocalizedtothegelsurface.Thenewdesigncanbeusedforintroducingbiologicalfunctionsintoself-assemblingpeptidestocreatescaffoldingmaterialswithpotentialapplicationsinareassuchastissueengineeringandregenerativemedicine.Ó2007ElsevierLtd.Allrightsreserved.
Keywords:Self-assembly;Peptide;Biomimeticmaterial;Matrixmetalloproteinase;Nanofiber;Hydrogel
1.IntroductionThenaturalextracellularmatrix(ECM)containsaplethoraofsignalsthatsynergisticallyactivatevariousintracellularsignalingpathwaystocontrolandguidethecellbehavior.RecapitulatingtheECMregulatorymechanismsisofcentralimportanceinfundamentalcellstudiesandcell-basedapplicationssuchastissueengineering.Methodshavebeenestablishedtocapturethenano-topographicalandbiochemi-calcharacteristicsinthenaturalECM[1,2].Mostofthesemethods,however,entailchemicalsynthesesthatlackanadequateflexibilitytogeneratetunablebiochemicalpatternsfortherationaldesignofextracellularenvironment.Inaddi-tion,athoroughandcomprehensiveunderstandingofECMecellinteractionsandthescreeningofbiomaterialsmayinvolvehigh-throughputstudiesthatrequiresystematicallyvaryingmaterialandbiochemicalpropertiesinafacileway[3,4].Weexplorehereapurepeptide-basedplatformfor*Correspondingauthor.Tel.:þ16172533107;fax:þ16172588827.
E-mailaddress:rlanger@mit.edu(R.Langer).
0142-9612/$-seefrontmatterÓ2007ElsevierLtd.Allrightsreserved.doi:10.1016/j.biomaterials.2007.11.046
Available online at www.sciencedirect.comBiomaterials29(2008)1713e1719www.elsevier.com/locate/biomaterialsenrichingbiologicalfunctionsinself-assemblingthree-dimensional(3-D)scaffolds.Anewfamilyofbiofunctionalmaterialscanpotentiallybeobtainedfromautomatedpeptidesynthesis.Self-assemblingpeptides(SAPs)haverecentlyemergedasanattractiveclassof3-Dscaffoldingmaterials,mainlyduetotheirnano-scalefibrousandporoustopographiesthatmimicthenaturalECMfeatures[5e8].Amongthem,peptides(arginine-alanine-aspartate-alanine)4((RADA)4)havebeen
usedtoformscaffoldsinsitufortissueengineeringapplica-tions[9e11].Thismaterialhasbeenshowntosupportthegrowthanddifferentiationofavarietyofcells,includingthoseoriginatedfromhuman,mammals,mouseandchicken,andcoveringstemcells,progenitorcellsandestablishedcelllines[12].Wereasonthatcellbehaviorcanbefurthercontrolledbycellematerialinteractionsifbiofunctionsaresynthesizedintothepeptidescaffolds.Thus,weinvestigatedanewSAPtemplatethatconsistsoftwomodules:oneforgeneratingorderedsecondarystructurestoenablehydrogelformation,andtheotherforsupplyingbiologicalcuestoelicitspecificcellularresponses.Cellularadhesionligands,suchasarginine-glycine-aspartate(RGD)fromfibronectinandtyrosine-isoleucine-glycine-serine-arginine(YIGSR)fromlaminin,havebeenwidelyusedinthetissue-scaffolddesigntoenhancecellattachmentandotherbasicfunctions[13].Inordertofurtherdirectcellbehavior,itisnecessarytoincorporateadditionalsignalsonthescaffoldtocommunicatewithcellsastheyremodeltheextracellularma-trix(ECM)[1].Morerecently,oligopeptidesthataresensitivetotheenzymaticcleavageofmatrixmetalloproteinases(MMPs)havebeenaddedtosyntheticpolymersandpeptide-amphiphiles[14,15].MMPsbelongtoafamilyofproteasesthatdegradeECMcomponentsandthereforeplayimportantrolesintissueregeneration.Theymakewayforcellstoexpand,allowtheECMtoberemodeled,andreleaseembeddedgrowthfactorsandothersignalsfromtheECMtostimulatecelldifferentiationandtissuegrowth[16,17].IncorporatingMMP-cleavablesubstratesintoSAPsisanattractivestrategytoengineeradynamicmechanismforelicitingcellandtissueremodelingactivities[1].TosynthesizeSAPspronetoremodelingbyECMproteases,weinsertedanMMP-2cleavablesequence,proline-valine-glycine-leucine-isoleucine-glycine(PVGLIG)intoanSAP.ThishexapeptidewasselectedfromscreeningacombinatorialpeptidelibraryforoptimalMMPsubstrates[18].ItsspecificityandsensitivitywaspreviouslydemonstratedinanMMP-sensitivepolymerepeptideedrugconjugate[19].TheoptimalplacementofPVGLIGwithinanSAPwasstudiedbysynthe-sizingaseriesofpeptideswithvariedPVGLIGpositioningandRADAblocklength.Theself-assemblingandgellingcapabilitiesoftheresultingmaterialswerecomparedandchar-acterizedbydynamicalmechanicalanalysis(DMA),circulardichroism(CD)spectroscopyandatomicforcemicroscopy(AFM).TheMMP-mediateddegradationoftheselectedPVGLIG-containingSAPwasstudiedbyafluorescencedetectionassay,massspectroscopicanalysisandnanoindenta-tionanalysis.